One confusing case for Mussa is when a single path between travelling between several sequences splits into more than one match on another sequence.
When one looks at the sequence alignment window it can look like some of the base pairs that were conserved for several species suddenly stop.
Most likely there's a thin red line leading off the screen which that base pair's match is actually part of.
The attached data provides a clearer example of this as the second match isn't that far away. However if one adds more Ns to sequence3 its not too hard to see how it looks like the base pair suddenly stops.
- Download and extract the example data.
- Create an analysis with sequence1.fa sequence2.fa and sequence3_close.fa with a threshold of 21 and a window size of 30.
- see how some of the base pairs connect to the right?
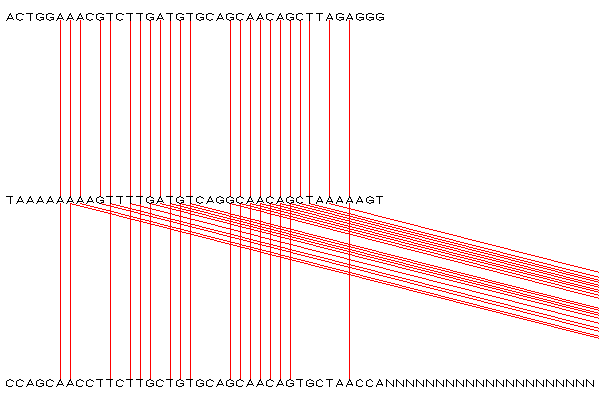
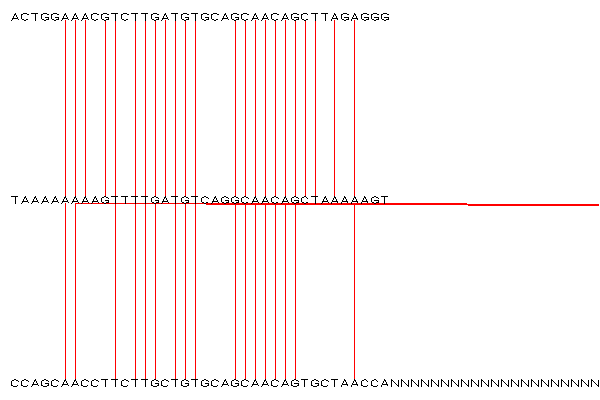
- Create a new analysis whith sequence3_close.fa replaced with sequence3_far.fa, and see how much harder it is to tell that a base pair is matching to a distant window.
Attachments (3)
-
overlapping_windows.zip
(905 bytes
) - added by 19 years ago.
Example data to illustrate how a distant match can be confusing
-
sequence_alignment_close.png
(6.7 KB
) - added by 19 years ago.
close windows
-
sequence_alignment_far.png
(3.4 KB
) - added by 19 years ago.
far away second window
Download all attachments as: .zip
